Build a Distributed Embedding Subsystem with MinIO, Langchain, and Ray Data

An embedding subsystem is one of four subsystems needed to implement Retrieval Augmented Generation. It turns your custom corpus into a database of vectors that can be searched for semantic meaning. The other subsystems are the data pipeline for creating your custom corpus, the retriever for querying the vector database to add more context to a user query, and finally, the serving subsystem that hosts your large language model (LLM) and will generate answers based on the user’s query and the context found in your vector database. The diagram below shows how these four subsystems work together during retrieval augmented generation.
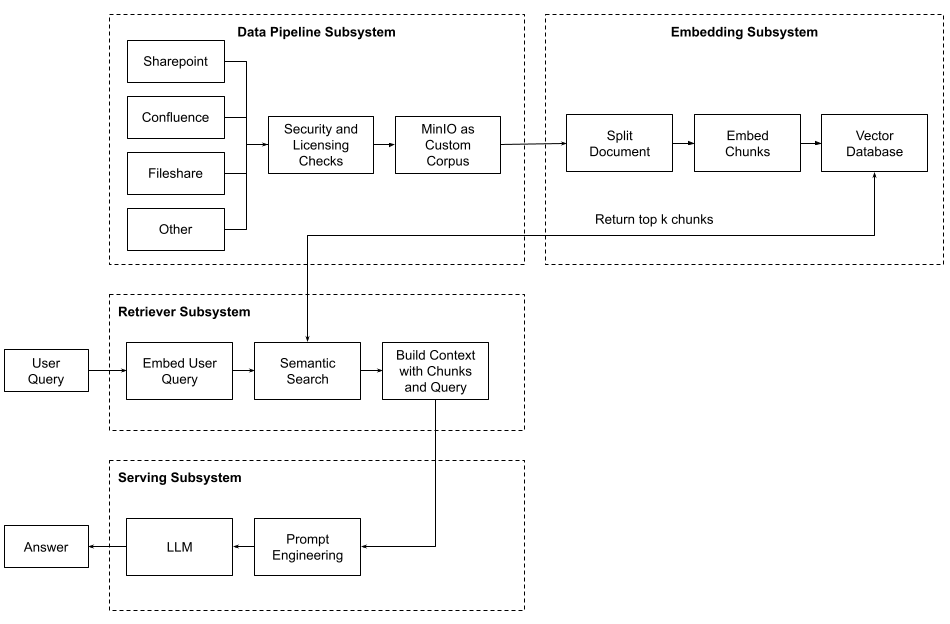
In this post, I want to focus on the embedding subsystem. In this subsystem, the documents that make up your organization's custom corpus are converted from their native format into text, broken up into smaller chunks, and then, for each chunk, an embedding is created (this is a vector usually with a dimension in the hundreds). Once the embedding is created, both the original chunk and the vector are stored in a vector database.
Embedding subsystems are conceptually easy to understand, and it is straightforward to implement a simple script that embeds a simple text file. However, if you have to implement an embedding subsystem for your organization, then how do you make the right design decisions for your organization, and how do you deal with the complexities that come with increasing demand? A few design decisions and real-world complexities are listed below:
- How to efficiently run multiple experiments to test different configuration options?
- How to handle tables and images in documents?
- How to deploy an embedding subsystem to a production environment?
- How to handle a high volume of documents that need to be embedded?
- What is the best vector database?
- What is the best storage option for documents, embedding models, and LLMs?
The first step to solving these issues is using modern tools capable of running on your engineering workstation as well as within a production environment. Specifically, we will use MinIO for all storage, Langchain for a low-code solution to document parsing (I’ll also provide some options that deal with images and tables better than Langchain.), and Ray Data to distribute both the chunking and embedding functions to a cluster. It should be no surprise that distributed technologies are the foundation of our solution. Not only do you get high throughput using commodity hardware setup for parallel processing, but the solution is cloud-native, making it portable across clouds and capable of running on-premise as well.
Let’s start by setting up a custom corpus for our experiments.
Setting up the Custom Corpus in MinIO
As illustrated above, the custom corpus is created by a data pipeline that aggregates documents that may be located in multiple portals across your organization into MinIO. Creating a document pipeline is a subject for another post - so for now, we will manually stage a few documents in a MinIO bucket. I will also only use text documents to keep things simple here. However, here are a couple of tips for dealing with multiple file formats and non-text within documents. First, look at Unstructured’s library for partitioning, cleaning, and extracting. Second, if you are dealing exclusively with PDFs, look at the Open-Parse library. We covered Open-Parse in a previous blog post, Improve RAG Performance with Open-Parse Intelligent Chunking.
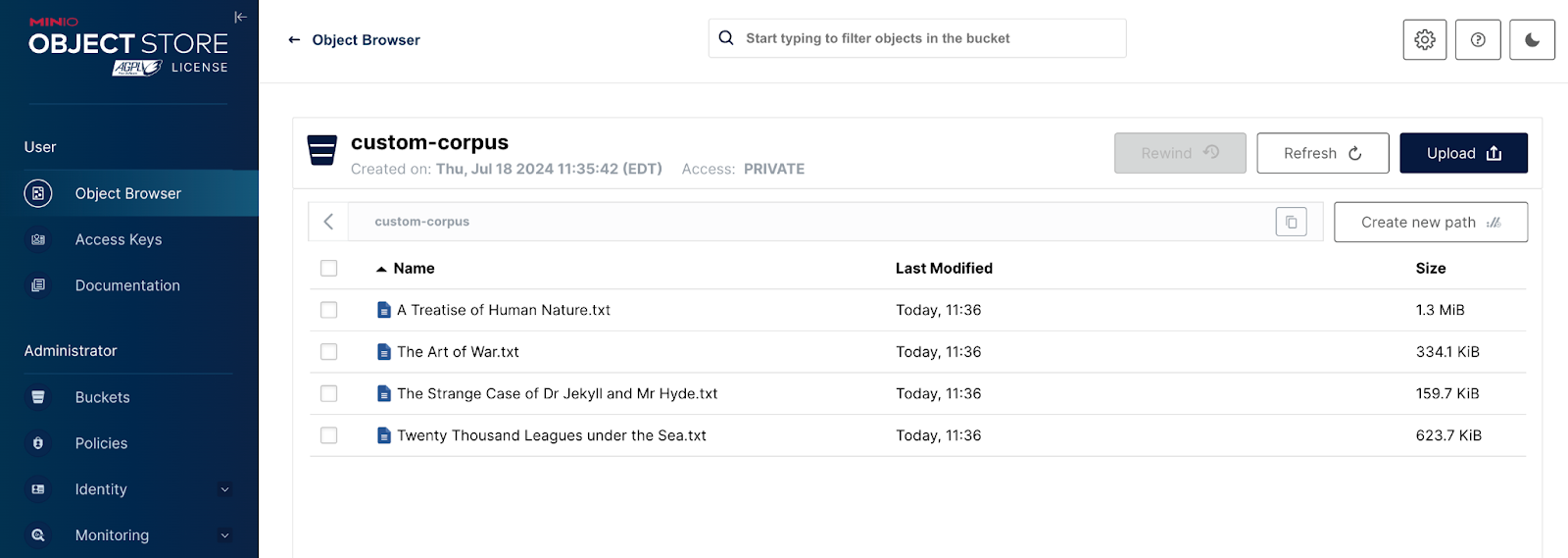
The screenshot below shows our custom corpus. I downloaded the text version of four popular books considered classics from Project Gutenberg.
- A Treatise of Human Nature - David Hume
- The Art of War - Sun Tzu
- The Strange Case of Dr Jekyll and Mr. Hyde - Robert Louis Stevenson
- Twenty Thousand Leagues Under the Sea - Jules Verne
Now that we have a custom corpus, we can set up a vector database to hold the embeddings.
Setting up MinIO and the Vector Database
The vector database I will use is Pgvector. Pgvector is an open-source extension for PostgreSQL that allows users to store, search, and analyze vector data within the database. The code download for this post has a docker-compose file that contains MinIO, Pgvector, and pgAdmin. Running the command below in the same directory as the docker-compose fill will bring up these three services as containers.
There is also an init.sql file (shown below). The docker-compose file maps this file to a startup directory for the container. This causes the SQL within the file to run, which creates the vector extension within Postgres and an “embeddings” table with the fields shown in the SQL file below.
Saving the Embedding Model to MinIO
The embedding model we will use is an open-source model from Hugging Face. The details are shown below. It is always a good idea to specify a specific version when running experiments.
Model Name: intfloat/multilingual-e5-smallRevision: ffdcc22a9a5c973ef0470385cef91e1ecb461d9f
Don’t be fooled by the model’s name. It is not small at all. It is 1.4GB. We need to download this model and upload it to MinIO. This is a one-time setup task to stage this model for experiments in a distributed environment. Unfortunately, the Hugging Face function we need (snapshot_download) does not have an S3 interface, so we will upload the model to MinIO using the MinIO Python SDK. A further complication is that a Hugging Face model is not a single file. It is a collection of files that get downloaded into a specified directory. We must upload this entire directory to MinIO, preserving the folder structure using MinIO paths. This is done using the “upload_model_to_minio” function shown below.
Running the following commands will use this function to upload our model to a bucket named “hf-models”.
The Embedding Function Library
When you use a library like Ray Data to distribute your data processing - which in this case is the chunking of text and the generation of embeddings for each chunk - all you are really doing is orchestrating simple function calls that perform one task in the process. All the functions we will need to create embeddings from a list of documents in a MinIO bucket are listed below, along with their parameters and return values. As you can see, we have everything we need to embed a collection of documents.
Creates a Python logger for sending debug, info, error, warning, and critical messages to a logging repository.
Downloads a model from MinIO to the current systems temp directory. It will delete it once it is loaded into memory.
Downloads a single document from MinIO and saves it to the current systems temp directory.
Returns a list of objects in the specified bucket. This list is sent to Ray Data, which distributes it evenly among all the Ray actors in the cluster.
Saves the embeddings along with the text chunks to the vector database.
Uploads the contents of the specified local directory to MinIO, preserving the folder structure as paths within the specified bucket.
Downloads a model from Hugging Face to the current systems temp directory and then uploads the model to the specified bucket, preserving folder structure as paths within the specified bucket.
A Simple Embedding Subsystem
Let’s use the functions above and create a simple non-distributed script. The code below will create embeddings for Robert Louis Stevenson's “The Strange Case of Dr Jekyll and Mr Hyde”.
First, we need to download the embedding model we wish to use and save it to MinIO. This is a one-time task; you do not need to do it every time you want to embed a new batch of models or run an experiment.
Next we need to download our model from MinIO, instantiate it, create a chunker (or splitter), create the embeddings and save them to our pgvector database.
Notice that we are using a GPU if we have access to one. Also, everything is configuration-driven, so running a different experiment is a matter of changing the configuration to reflect the experiment you wish to run. This includes changing the embedding model if you wish.
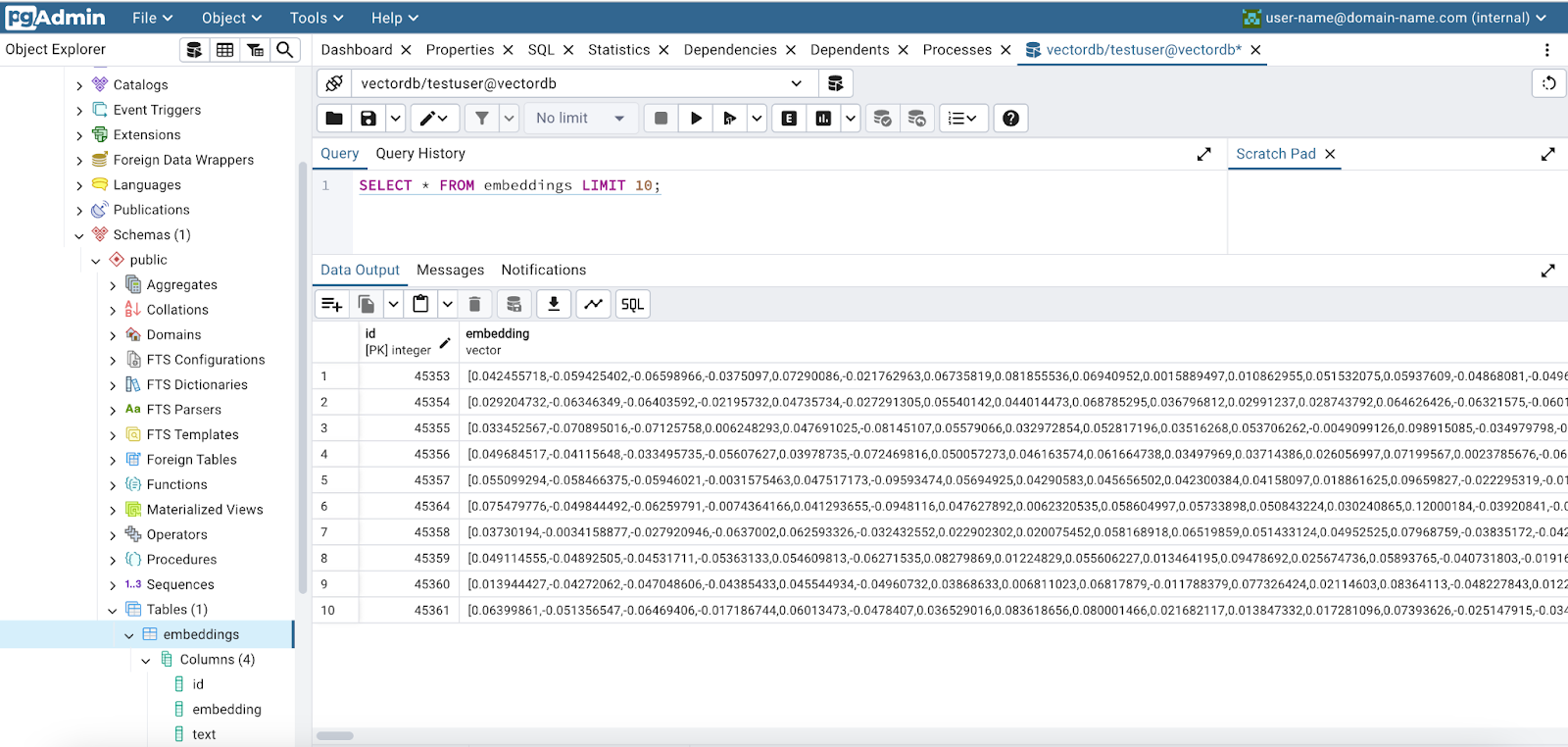
Below is a screenshot of pgAdmin that shows our newly created embeddings.
Now that we have a simple script that creates embeddings for a single document, the next step is to migrate this code to a framework that runs in a cluster. This will allow an entire corpus of documents to be embedded in parallel. We will use Ray Data to do this.
Distributing the Embedding Subsystem
The first step in distributing our embedding subsystem is to put all the work into a class that behaves like a function. This is done using Python’s “__call__” built-in method. (This is a Ray Data requirement.) Our class is shown below.
Ray Data will instantiate this class once for each actor in the Ray cluster we will create shortly. This object will stay alive and receive multiple batches to process. Notice that the “__init__” function is downloading our embedding model, creating a SentenceTransformer with it. The SentenceTransformer class makes it easy to use embedding models. Additionally we are using the RecursiveCharacterTextSplitter from LangChain to split or chunk our documents. It recursively splits text based on a list of characters (we are using its default list), starting with the first character in the list and moving on to the next if the first split is too large. The goal is to keep related pieces of text together, preserving their semantic relationship. All this setup work happens only once when the Embed object is created. We could have used a simple function for distributing the work, but this setup work would have to get done for each batch, and that is not the correct design when you have setup work to do.
Next, we need to initialize the Ray cluster.
In our demo here we are creating a local Ray instance. I am not using a Kubernetes cluster. This is the best way to get your code working before moving to a real cluster. We are not creating any Ray actors yet - but we are sending Ray configuration information that tells Ray the environment variables and libraries that are needed for each actor.
Next we create a Ray dataset to hold all the data we want to send to the instances of our Embed class that will be running within each Ray actor. In our case, each Ray actor is going to receive a list of object references (the path to the documents) that are stored in MinIO. We will use the “get_object_list” function from our function library described above. The Ray dataset that is returned from “ray.data.from_items() contains logic that will turn this list into smaller batches for each actor when we kick off the distributed embedding process.
We are almost ready to do some distributed computing, but we have one more coding task to complete. We need to map our Ray dataset to our Embed class and tell Ray how to set up the cluster we previously initialized for this workload. This is done using the Ray dataset's “map_batches” method. You can send a function or a callable class to “map_batches.” If you send a function, Ray Data uses stateless Ray tasks. For classes, Ray Data uses stateful Ray actors.
Notice that we are passing in the Embed class that needs to be instantiated for each actor. We are also specifying the number of actors, the batch size for each call to an actor, and finally the number of GPUs and CPUs accessible from each actor. The map_batches method returns another Ray dataset (ds_embed) that contains all the return values from all the actors. This is a collection of return values from the “__call__” method in Embed.
Finally, we are ready to kick off our distributed embedding job. You may have noticed that the previous command ran very quickly. That is because no computations have happened yet. Transformations in Ray (map_batch is considered a transformation) are “lazy.” They are not executed until you trigger consumption of the data by iterating over the dataset, saving the dataset, or inspecting the properties of the dataset. So, we need to ask ds_embed for the return values from our actors. This is done below. The snippet below will take some time to run.
That is it. We are done. Once the code above completes you will see output similar to what is shown below.
Summary
In this post, we built a distributed embedding subsystem that can run on engineering workstations and in a fully distributed cloud-native production environment. The code presented has the following benefits that directly address the complexities and real-world concerns identified in our introduction.
- Experiments can be run efficiently, allowing the different configuration options to be thoroughly tested.
- In addition to configuration options, parsing options should be experimented with. This will allow you to handle multiple file types and deal with non-text data within a document.
- When using the code shown here, your production environment will run the same code engineers use for testing and experimenting.
- A distributed embedding subsystem can run in a cluster. Clusters can quickly scale to handle a large number of documents that need to be processed as a batch and can also be scaled for real-time workloads.
- The code presented in this post encapsulates vector database calls, allowing engineers to swap in different products.
- MinIO is the best storage solution for generative AI. As we saw in this post, the embedding models and the documents must be stored in a high-speed, scalable storage solution.
If you have any questions, be sure to reach out to us on Slack!






